Table of Contents
Joshua Akey
Princeton University
In Joshua’s lab, his work is guided by the principle that answers to complex and important problems should not be confined by rigid disciplinary boundaries.

When the first modern humans arose in East Africa sometime between 200,000 and 300,000 years ago, the world was very different compared to today. Perhaps the biggest difference was that we – meaning people of our species, Homo sapiens – were only one of several types of humans (or hominins) that simultaneously existed on Earth.
From the well-known Neanderthals and more enigmatic Denisovans in Eurasia, to the diminutive “hobbit” Homo floresiensis on the island of Flores in Indonesia, to Homo naledi that lived in South Africa, multiple hominins abounded.
Then, between 30,000 and 40,000 years ago, all but one type of these hominins disappeared, and for the first time we were alone.
Until recently, one of the mysteries about human history was whether our ancestors interacted and mated with these other types of humans before they went extinct. This fascinating question was the subject of great and often contentious debates among scientists for decades, because the data needed to answer this question simply didn’t exist. In fact, it seemed to many that the data would never exist.
Svante Pääbo, however, paid little attention to what people thought was or was not possible. His persistence in developing tools to extract, sequence and interpret ancient DNA enabled sequencing the genomes of Neanderthals, Denisovans and early modern humans who lived over 45,000 years ago.
For developing this new field of paleogenomics, Pääbo was awarded the 2022 Nobel Prize in Physiology or Medicine. This honor is not only well-deserved recognition for Pääbo’s triumphs, but also for evolutionary genomics and the insights it can contribute toward a more comprehensive understanding of human health and disease.
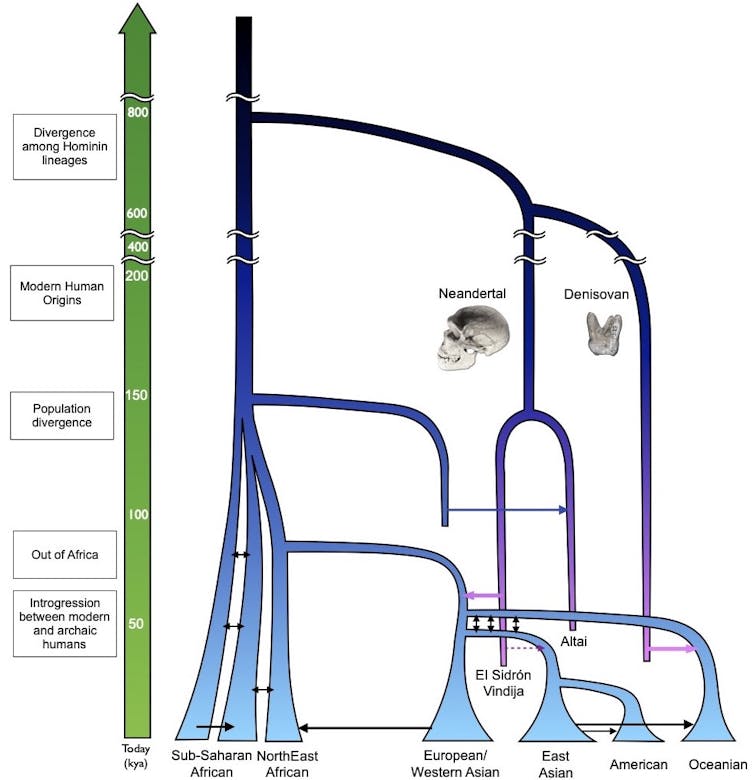
Mixing and mating, revealed by DNA
Genetic studies of living people over the past several decades revealed the general contours of human history. Our species arose in Africa, dispersing out from that continent around 60,000 years ago, ultimately spreading to nearly all habitable places on Earth. Other types of humans existed as modern humans migrated throughout the world, but the genetic data showed little evidence that modern humans mated with other hominins.
Over the past decade, however, the study of ancient DNA, recovered from fossils up to around 400,000 years old, has revealed startling new twists and turns in the story of human history.
For example, the Neanderthal genome provided the data necessary to definitively show that humans and Neanderthals mated. Non-African people alive today inherited about 2% of their genomes from Neanderthal ancestors, thanks to this kind of interbreeding.
In one of the biggest surprises, when Pääbo and his colleagues sequenced ancient DNA obtained from a small finger bone fragment that was assumed to be Neanderthal, it turned out to be an entirely unknown type of human, now called Denisovans. Humans and Denisovans also mated, with the highest levels of Denisovan ancestry present today – between 4% and 6% – in individuals of Oceanic ancestry.
Strikingly, ancient DNA from a 90,000-year-old female revealed that she had a Neanderthal mother and a Denisovan father. Although there are still many unanswered questions, the picture emerging from analyses of ancient and modern DNA is that not only did multiple hominins overlap in time and space, but that matings were relatively common.

Archaic genes you carry today
Estimating the proportion of ancestry that modern individuals have from Neanderthals or Denisovans is certainly interesting. But ancestry proportions provide limited information about the consequences of these ancient matings.
For instance, does DNA inherited from Neanderthals and Denisovans influence biological functions that occur within our cells? Does this DNA influence traits like eye color or susceptibility to disease? Were DNA sequences from our evolutionary cousins ever beneficial, helping humans adapt to new environments?
To answer these questions, we need to identify the bits of Neanderthal and Denisovan DNA scattered throughout the genomes of modern individuals.
In 2014, my group and David Reich’s group independently published the first maps of Neanderthal sequences that survive in the DNA of modern humans. Today, roughly 40% of the Neanderthal genome has been recovered not by sequencing ancient DNA recovered from a fossil, but indirectly by piecing together the Neanderthal sequences that persist in the genomes of contemporary individuals.
Similarly, in 2016 my group and David Reich’s group published the first comprehensive catalogs of DNA sequences in modern individuals inherited from Denisovan ancestors. Surprisingly, when we analyzed the Denisovan sequences that persist in people today, we discovered they came from two distinct Denisovan populations, and therefore at least two separate waves of matings occurred between Denisovans and modern humans.
The analysis of Neanderthal and Denisovan DNA in modern humans reveals that some of their sequence was harmful and rapidly got purged from human genomes. In fact, the initial fraction of Neanderthal ancestry in humans who lived approximately 45,000 years ago was around 10%. That amount rapidly declined over a small number of generations to the 2% observed in contemporary individuals.
The removal of deleterious archaic sequences also created large regions of the human genome that are significantly depleted of both Neanderthal and Denisovan ancestry. These deserts of archaic hominin sequences are interesting because they may help identify genetic changes that contribute to uniquely modern human traits, such as our capacity for language, symbolic thought and culture, although there is debate about just how unique these traits are to modern humans.
In contrast, there are also sequences inherited from Neanderthals and Denisovans that were advantageous, and helped modern humans adapt to new environments as they dispersed out of Africa. Neanderthal versions of several immune-related genes have risen to high frequency in several non-African populations, which likely helped humans fend off exposure to new pathogens. Similarly, a version of the EPAS1 gene, which contributes to high-altitude adaptation in Tibetan populations, was inherited from Denisovans.
It is also becoming clear that DNA sequences inherited from Neanderthal and Denisovan ancestors contribute to the burden of disease in present day individuals. Neanderthal sequences have been shown to influence both susceptibility to and protection against severe COVID-19. Archaic hominin sequences have also been shown to influence susceptibility to depression, Type 2 diabetes and celiac disease among others. Ongoing studies will undoubtedly reveal more about how Neanderthal and Denisovan ancestry contributes to human disease.

I was a graduate student when the Human Genome Project was nearing completion a little over two decades ago. I was drawn to genetics because I found it fascinating that, by analyzing the DNA of present-day individuals, you could learn aspects about a population’s history that occurred tens of thousands of years ago.
Today, I am just as fascinated by the stories contained in our DNA, and the work of Svante Pääbo and his colleagues has enabled these stories to be told in a way that simply was not possible before.
This article is republished from The Conversation under a Creative Commons license. Read the original article.